lrgasp-submissions
Submission structure
Submissions to LRGASP consist of a set of experiments targeting a particular challenge submitted by a team. With two types of experiments: model and expression. Model experiments are genomic transcript model predictions, and expression experiments are transcript expression quantification of a set of submitted models.
A given challenge may combine results from multiple, distinct experiments. For instance, the high-quality genome isoform annotation challenge will include both human and mouse experiments. To accommodate this, experiments are grouped into entries against a particular challenge. An entry is the unit of both submission and evaluation. The experiments in an entry should uses as similar a set of parameters and data to allow for the meaningful combination of the results.
Symbolic experiment identifiers assigned by the team and must be valid symbolic identifier, as described below.
The example entries will be useful in understanding structure.
Submission overview
This diagram shows the general logical and directory structure of LRGASP entries, which are explain below.
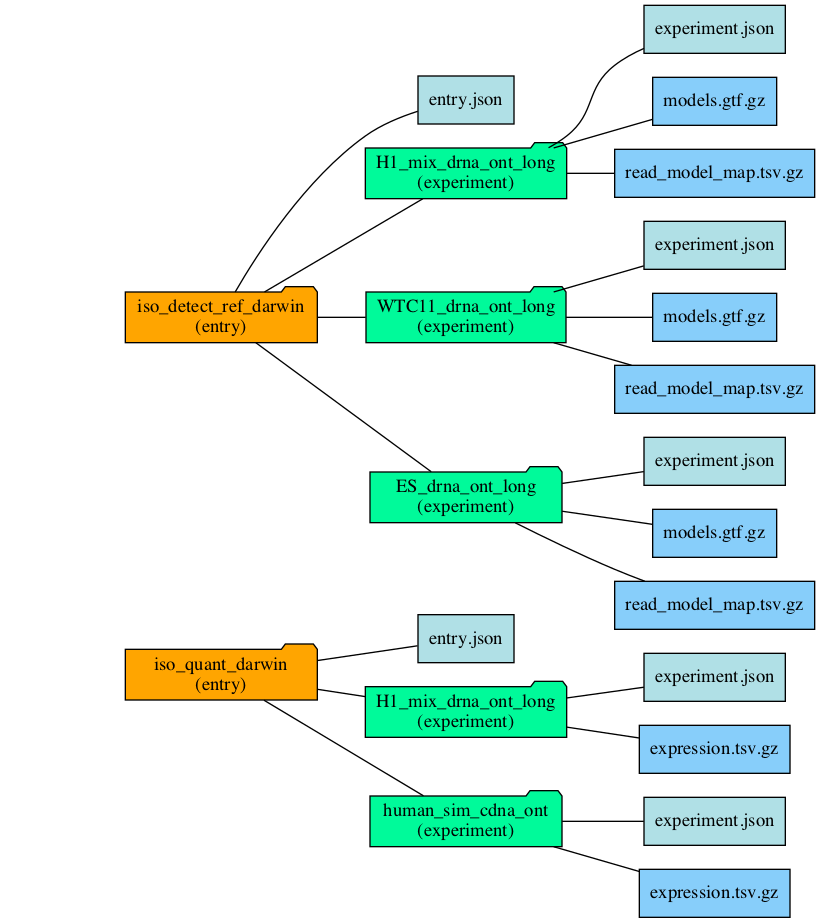
Entry structure
Each entry must conform to fix, straight-forward file hierarchy.
Each entry to a challenge is in a directory with the same name as the entry_id.
The entry ids must be prefixed with the Challenge identifier followed by
a _ and a unique team-defined name. For instance:
iso_detect_ref_darwin_drna_ontiso_quant_ont_darwiniso_detect_de_novo_darwin
The entry directory contains an entry.json entry metadata
describing the entry. There is a directory per experiment within the entry directory, with each directory named the same as the
submitter-defined ``experiment_id`.
All entries MUST be validated with the provided submission tools before uploading.
Experiment structure
Challenge 1 model experiments must contain the following files:
experiment.json- Experiment metadata describing the experiment resultsmodels.gtf.gz- GTF file with model annotations, compressed with gzip.read_model_map.tsv.gzRead to model map file that associates every transcript model the GTF with a least one read.
Challenge 2 quantification experiments must contain the following files:
experiment.json- Experiment metadata describing the experiment resultsexpression.tsv.gz- Expression matrix file with the results of the experiment, which must be in TPM.models.gtf.gz- GTF file with target model annotations, compressed with gzip.
Challenge 3 novel model experiments must contain the following files:
experiment.json- Experiment metadata describing the experiment resultsrna.fasta.gz- FASTA file with model RNA sequences, compressed with gzip.read_model_map.tsv.gzRead to model map file that associates every transcript model in the FASTA with a least one read.